Regulation of Type VI secretion system gene clusters
The Type VI secretion gene clusters are subjected to a broad variety of transcriptional, post-transcriptional and post-translational regulatory mechanisms. For examples, the Pseudomonas aeruginosa H1-T6SS is regulated by the RetS/GacAS/RsmAYZ pathway which involves two-component regulation system and small RNA, the enteroaggregative E. coli sci-2 gene cluster is regulated by a transcriptional regulator of the AraC family, while several other are under the control of Quorum sensing or alternative sigma factors. Repression of T6SS genes by histone-like proteins has also been reported. For a comprehensive list of the regulatory mechanisms underlying T6SS gene expression, we refer to recent reviews: Bernard et al., 2010; Leung et al., 2011; Silverman et al., 2012; and Miyata et al., 2013.
In the recent years, we have been interested in deciphering the regulatory mechanisms for a subset of T6SS gene clusters, including the EAEC sci-1 or the T6SS-4 gene cluster of Yersinia pseudotuberculosis.
EAEC sci1
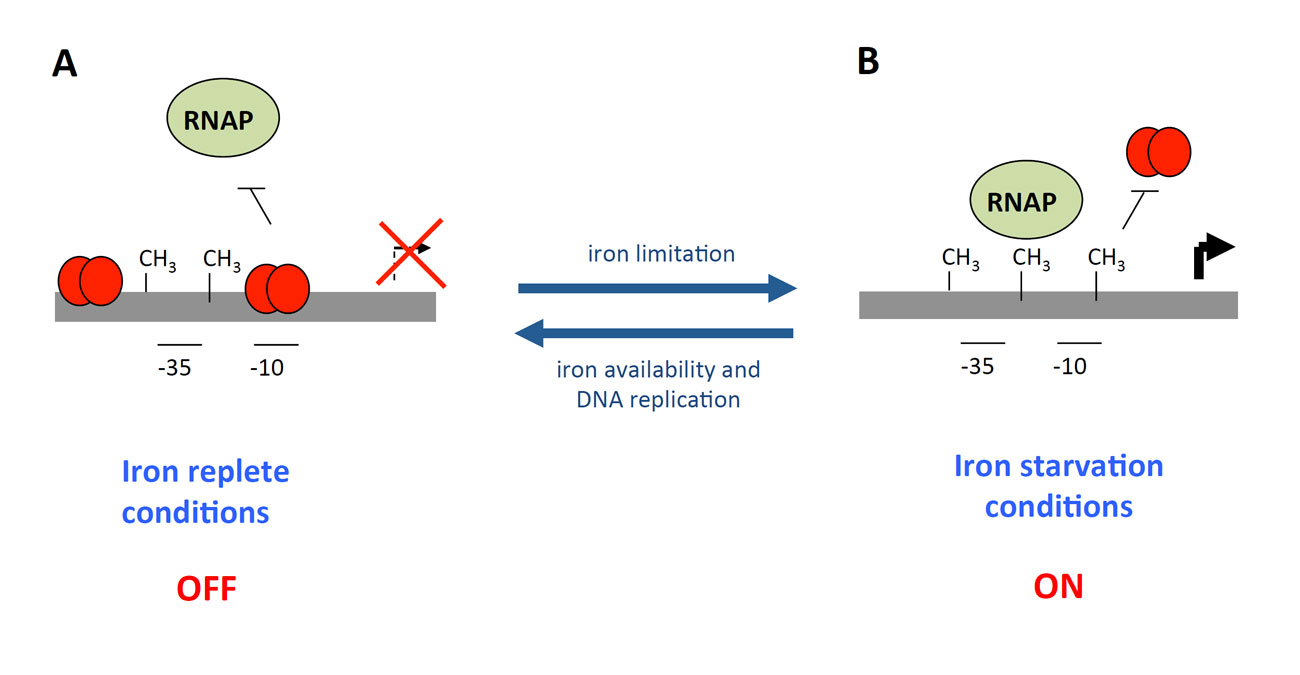
By random transposon mutagenesis of an EAEC strain bearing a transcriptional sci1-lacZ reporter fusion, Yannick identified the Fur repressor as a regulator of the sci-1 T6SS gene cluster. The implication of Fur in the regulatory mechanism was confirmed by the clean deletion of the fur gene, its complementation and by the increased activity of the reporter fusion after iron depletion by the adition of the iron chelator 2,2’-dipyridyl. The sci-1 promoter region possesses two typical Fur-binding site (Fur1 and Fur2), with the Fur1 site overlapping with the -10 of transcription. These motifs were confirmed as Fur-binding boxes by in vitro gel shift and by in vivo Fur titration experiments. However, while this mechanism appears quite simple at first, we also found that the sci-1 promoter region carries three Dam methylation sites, GATC-I to –III, and that the Fur1 box overlaps with GATC-I.
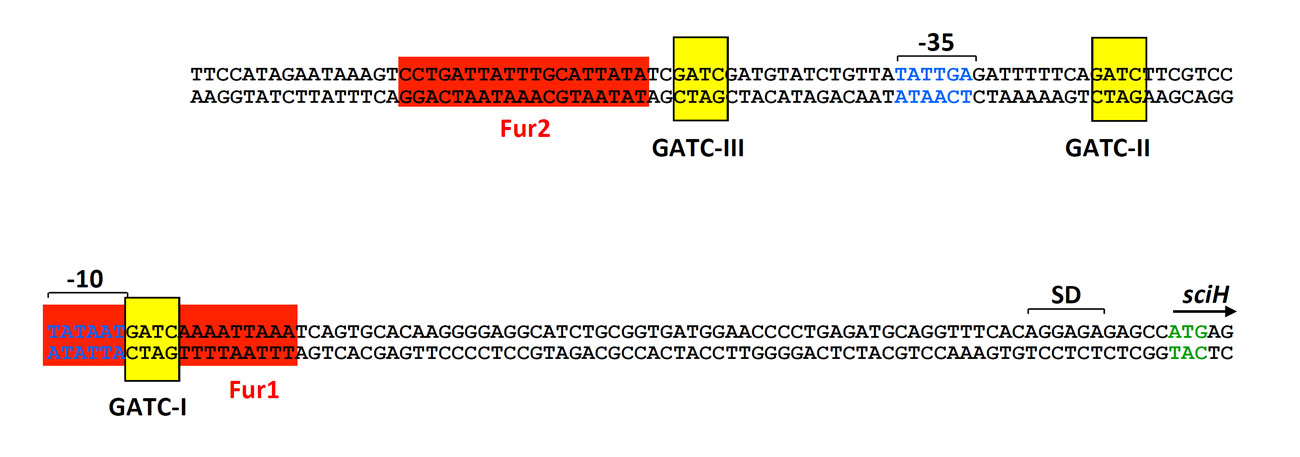
Promoter sequence of the sci-1 T6SS gene cluster. The position of the -10 and -35 is shown in blue letters. The Fur boxes and the Dam methylation sites are highlighted in red and yellow respectively.
Indeed, Yannick found that these sites are targets for the Dam methylase in vitro and in vivo. In vivo, the GATC-1 is not methylated; however, in the fur mutant strain or in presence of 2,2’-dipyridyl, this site is methylated. Therefore Fur binding prevents methylation of GATC-I. By contrast, methylation of GATC-I decreases the affinity of Fur for the methylated Fur1 box. Therefore Fur and Dam have antagonistic role, allowing the fine-tuned regulation of the cluster. In the presence of iron, the sci-1 gene cluster is repressed by Fur. When iron is limiting, Fur is relieved from the promoter, allowing its expression. However, if the conditions are favourable, replication occurs, as well as methylation, and the methylation of GATC-I site allows the constitutive ON expression of the cluster (Brunet et al., 2011). This mechanism is similar to those reported for the regulation of ag43 or of the pap operon, although the transcriptional regulators are different.
Sigma-54 and Enhancer Binding Proteins
While exploring whether other T6SS promoters contain Fur-binding motifs and/or Dam methylation sites, we observed that several of them, mostly in environmental bacteria (Pseudomonas syringae, Xanthomonas, Ralstonia, Pectobacterium, Vibrio cholerae, Aeromonas hydrophila, Marinomonas,…), carry σ54 specific boxes. By contrast to other sigma factors, σ54 requires the action of a dedicated transcriptional regulator of the bEBP (bacterial Enhancer Binding Proteins) family. Interestingly, bEBP-encoding genes are found in the T6SS gene clusters bearing the σ54 boxes. By using a combination of transcriptional fusions in the E. coli heterologous host and electrophoretic mobility shift assays, we reported that the T6SS gene clusters of Vibrio cholerae, Aeromonas hydrophila, Pectobacterium atrosepticum, Marinomonas sp. and Pseudomonas syringae pv. tomato are indeed under the control of the σ54 factors and their cognate bEBP (Bernard et al., 2011).
By using the same approaches, we participated to the characterization of a regulatory mechanism of an gene operon in Desulfovibrio vulgaris that involves σ54, a dedicated bEBP and IHF. These studies were developed by Anouchka Fiévet and Corinne Aubert in the team of Alain Dolla (UMR7283) and published in Journal of Bacteriology and PLOS ONE (Fiévet et al. 2011; Fiévet et al., 2014).
Yersinia pseudotuberculosis T6SS-4
Yersinia pseudotuberculosis is an interesting bacterium regarding the Type VI secretion system, as five complete and one incomplete T6SS gene clusters are encoded on the genome. The T6SS-4 cluster was previously shown to be thermoregulated, suggesting it has a role during the infection cycle of Yersinia. By random mutagenesis, Erwan found several regulators, including proteins that participate to the cell envelope biogenesis and the EnvZ-OmpR two-component regulator, suggesting that the expression of this cluster is responsive to a cell envelope stress and/or osmolarity. We also showed that this cluster is upregulated in presence of bile salts such as deoxycholate. The purification of OmpR further showed that this regulator directly binds to the promoter region of T6SS-4, in agreement with the presence of several OmpR-like binding boxes (Gueguen et al., 2013). Similar results have been published by Zhang and co-workers (Zhang et al., 2013).
Salmonella enterica Typhimurium
By using transcriptional reporter fusions, electrophoretic mobility shift assays and DNase footprints, we have shown in collaboration with the group of Sylvie Rimsky (ENS Cachan) that the T6SS gene cluster encoding on the SPI-6 genetic island of Salmonella enterica sv. Typhimurium is repressed by the histone-like nucleoid-associated protein H-NS. This confirmed previous genome-wide micro-arrays and ChIP-on-chip studies from the Jay Hinton’s and Ferric Fang’s laboratories (Navarre et al., 2006 ; Lucchini et al., 2006). We further demonstrated that the Salmonella T6SS has an anti-bacterial activity (Brunet et al., 2015).
Transcriptional slippage in Citrobacter rodentium
Erwan initiated a study on the Citrobacter rodentium T6SS, a gene cluster which is silent in the laboratory conditions. Although he failed to identify the conditions and regulator(s) required for the activation of the expression of this cluster, he reported that swapping the divergent promoter of the cluster with inducible promoters activates its expression. Further studies showed that this cluster, once activated, is functional and confers anti-bacterial activity (Gueguen et al., 2013).
However, this result was intriguing as the C. rodentium tssM gene encodes a truncated protein, lacking the C-terminal domain found in canonical TssM proteins and mediating the interaction with the TssJ lipoprotein. However, we noticed that the stop codon is preceded by a run of 11 consecutive adenosines. In collaboration with the group of John Atkins (Salt Lake City, USA) and by using a combination of Western-blot, GFP fluorescence and mass spectrometry analyses, Erwan demonstrated that this poly-A tract is a transcriptional slippery site that causes the incorporation of additional adenosines into the mRNA during transcription, leading to frameshifting and hence the production of two TssM1 variants including a full-length canonical protein. Finally, he showed that both forms of TssM1 are required for the function of the T6SS in C. rodentium. The frequency of frameshifting is 20–25% and therefore yields a molecular ratio of 3–4:1 between the truncated and the full-length variants. This ratio between the two forms is critical as inverting this ratio leads to a non-functional T6SS apparatus. Finally, Erwan showed that a similar frameshiting mechanism occurs in the tssM gene associated with the Yersinia pseudotuberculosis T6SS-3 gene cluster (Gueguen et al., 2014).
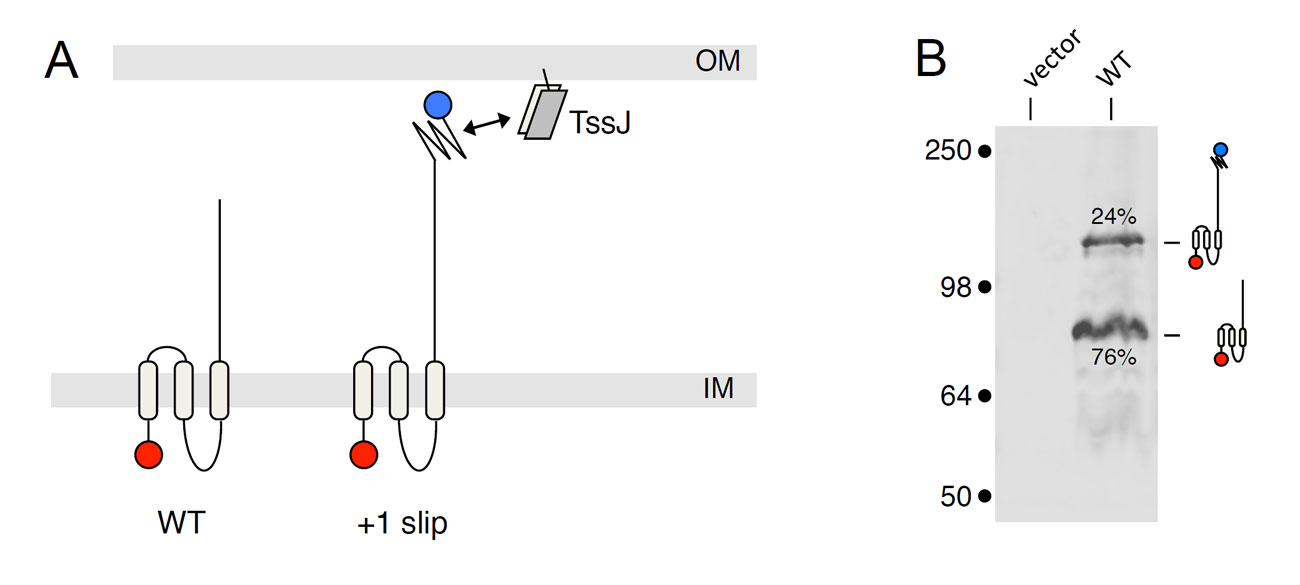
Transcriptional slippery in the tssM gene of Citrobacter rodentium. (A) Schematic representation of the TssM variants obtained by normal transcription (WT) and +1 slip. The full-length TssM protein produced in case of +1 slip carries the C-terminal domain that contacts the TssJ outer membrane lipoprotein. (B) Western-blot analyses showing that two forms are produced in vivo, with their relative ratio.